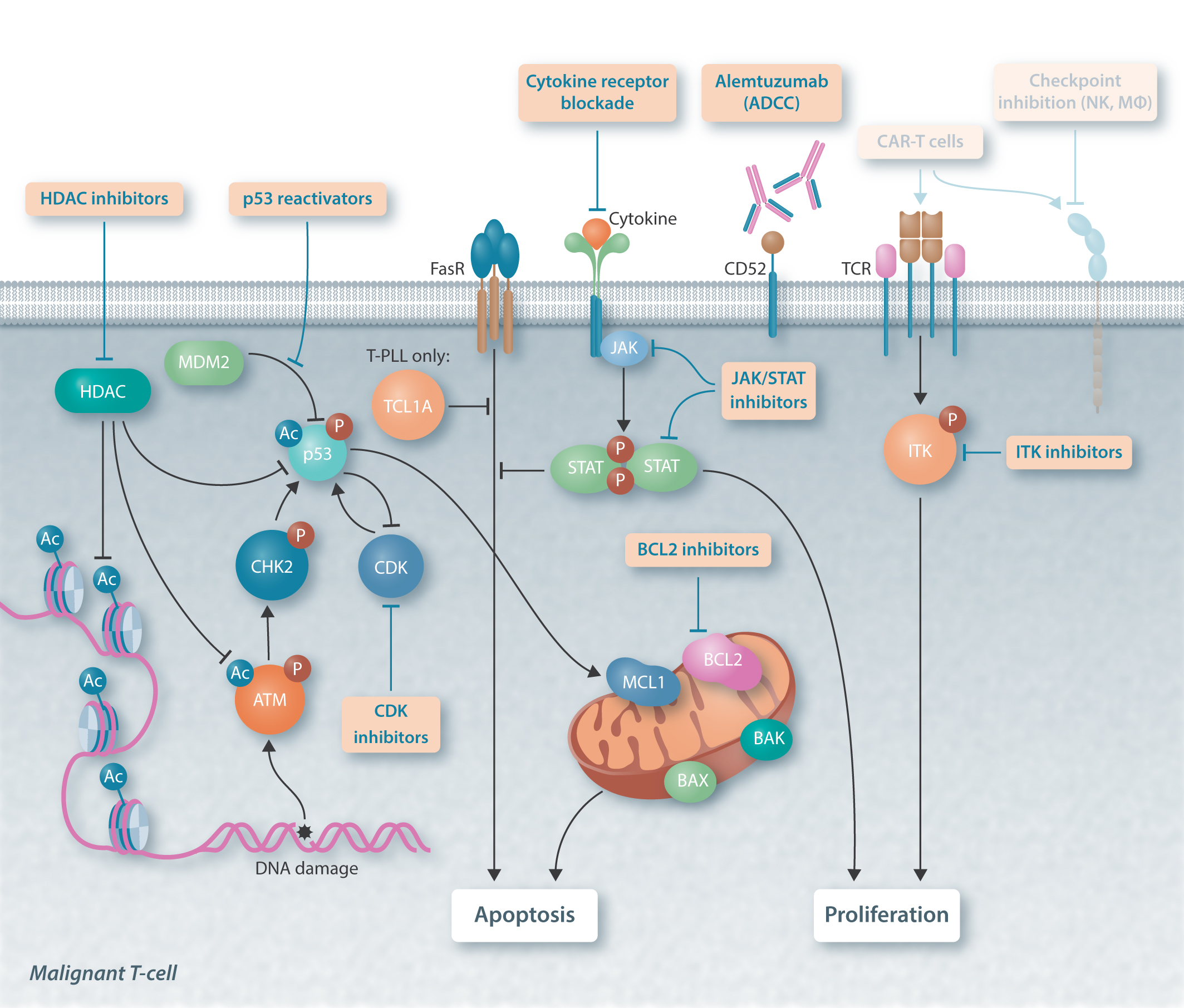
JAK/STAT-Activating Genomic Alterations Are a Hallmark of T-PLL
Missense mutations of JAK and STAT genes are frequently found in T-PLL. However, sequencing analyses of the JAK/STAT pathway have been performed in small cohorts, due to the limited number of patient samples. Here we conducted a meta-analysis that re-evaluated the genomic landscape of the JAK/STAT pathway and its regulators in 275 T-PLL cases. Interestingly, next to gain of function mutations in JAK and STAT genes, a significant proportion of genes encoding for potential negative regulators of STAT5B showed genomic losses. Overall, considering such losses of negative regulators and the GOF mutations in JAK and STAT genes, we found that a total of 89.8% of T-PLL revealed a genomic aberration potentially explaining enhanced STAT5B activity.
Read the paper:
JAK/STAT-Activating Genomic Alterations Are a Hallmark of T-PLL